Research
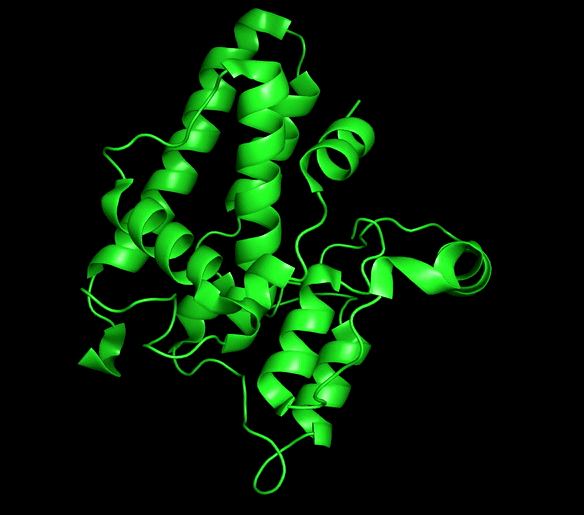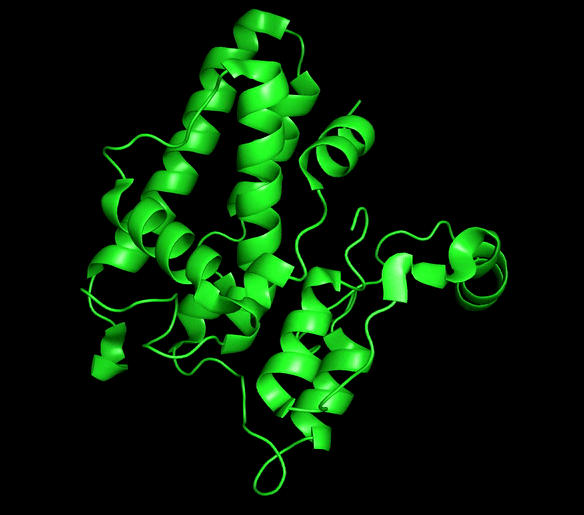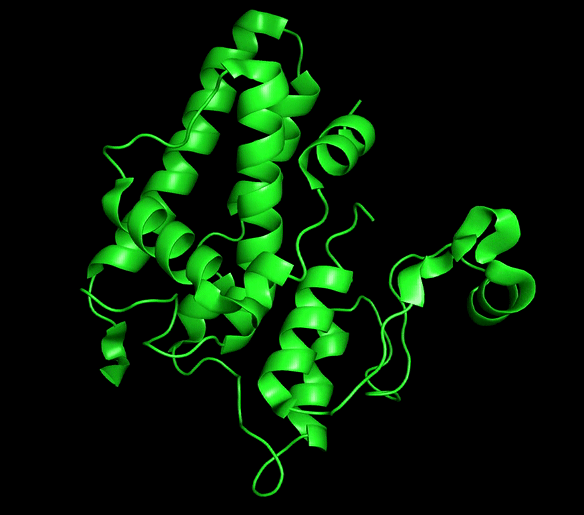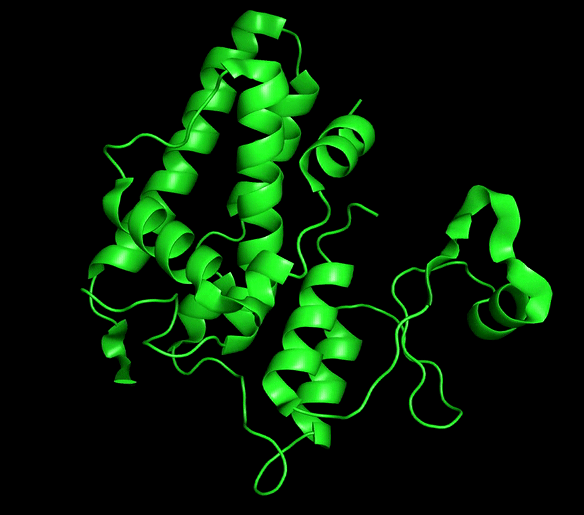
Chloride ion channels control several cellular processes required for the normal function of the cell, and growing evidence supports the role of this channels in the development of different types of cancer. The Chloride intracellular channel (CLIC) family is a recently identified class of chloride ion channel proteins that can exist both as cytoplasmic soluble proteins and as integral membrane elements with ion channel activity. This unusual equilibrium is regulated by reactive oxidative species (ROS) and pH changes. However, little is known about the structural transition between the two forms.
CLIC1 and CLIC4 have been directly implicated in tumor development and identified as novel therapeutic targets. Our main aim is to obtain mechanistic information for this process that can be of great use for the development of conformation-specific pharmacological inhibitors and regulators that could lead to new avenues for cancer treatment.
To this end, we use solution state NMR as the main tool to obtain mechanistic information of the membrane insertion process with atomic detail in combination with Fluorescence microscopy, Circular dichroism and other biophysical tools.
In addition, we use a combination of solution NMR and X-ray crystallography to gain insights into the structure of the channels and their regulation by different ligands.
Chloride intracellular channels (CLICs)
-
Ossa, F., Schnell, J. R. & Ortega-Roldan J.L. (2016). Sigma-1 Receptor Structure. Chapter of the book: Sigma Receptors: Their Role in Disease and as Therapeutic Targets. Springer’s Advances in Experimental Medicine and Biology series.
-
Ortega-Roldan, J. L*., Ossa, F., Amin, N. & Schnell, J. R*. (2015). Solution NMR studies reveal the location of the second transmembrane domain of the human sigma-1 receptor. FEBS Letters. doi:10.1016/j.febslet.2015.01.033
-
Ortega-Roldan, J. L., Ossa, F., & Schnell, J. R*. (2013). Characterization of the human Sigma-1 Receptor chaperone domain structure and BiP interactions. The Journal of Biological Chemistry. doi:10.1074/jbc.M113.450379
-
Ortega Roldan, J. L., Casares, S., Ringkjøbing Jensen, M., Cárdenes, N., Bravo, J., Blackledge, M., et al. (2013). Distinct ubiquitin binding modes exhibited by SH3 domains: molecular determinants and functional implications. PloS One, 8(9), e73018. doi:10.1371/journal.pone.0073018
-
Ceregido, M. A., Garcia-Pino, A., Ortega Roldan, J. L., Casares, S., Lopez Mayorga, O., Bravo, J., et al. (2013). Multimeric and differential binding of CIN85/CD2AP with two atypical proline-rich sequences from CD2 and Cbl-b*. The FEBS Journal, 280(14), 3399–3415. doi:10.1111/febs.12333
-
Guerry, P. P., Salmon, L. L., Mollica, L. L., Ortega-Roldan, J.L. J. O., Markwick, P. P., van Nuland, N. A. J. N., et al. (2013). Mapping the population of protein conformational energy sub-States from NMR dipolar couplings. Angewandte Chemie (International Ed. in English), 52(11), 3181–3185. doi:10.1002/anie.201209669
-
Salmon, L., Pierce, L., Grimm, A., Ortega-Roldan, J. L., Mollica, L., Jensen, M. R., et al. (2012). Multi-Timescale Conformational Dynamics of the SH3 Domain of CD2-Associated Protein using NMR Spectroscopy and Accelerated Molecular Dynamics. Angewandte Chemie (International Ed. in English), 51(25), 6103–6106. doi:10.1002/anie.201202026
-
Ortega-Roldan, J. L. O., Blackledge, M., van Nuland, N. A. J., & Azuaga, A. I. (2011). Solution structure, dynamics and thermodynamics of the three SH3 domains of CD2AP. Journal of Biomolecular NMR, 50(2), 103–117. doi:10.1007/s10858-011-9505-5
-
Salmon, L., Ortega-Roldan, J. L., Lescop, E., Licinio, A., van Nuland, N., Jensen, M. R., & Blackledge, M. (2011). Structure, dynamics, and kinetics of weak protein-protein complexes from NMR spin relaxation measurements of titrated solutions. Angewandte Chemie (International Ed. in English), 50(16), 3755–3759. doi:10.1002/anie.201100310
-
Jensen, M. R., Ortega-Roldan, J. L., Salmon, L., van Nuland, N., & Blackledge, M. (2011). Characterizing weak protein-protein complexes by NMR residual dipolar couplings. European Biophysics Journal : EBJ, 40(12), 1371–1381. doi:10.1007/s00249-011-0720-5
-
Ortega-Roldan, J. L., Jensen, M. R., Brutscher, B., Azuaga, A. I., Blackledge, M., & van Nuland, N. A. J. (2009). Accurate characterization of weak macromolecular interactions by titration of NMR residual dipolar couplings: application to the CD2AP SH3-C:ubiquitin complex. Nucleic Acids Research, 37(9), e70. doi:10.1093/nar/gkp211
-
Ortega Roldan, J. L., Romero Romero, M. L., Ora, A., Ab, E., Lopez Mayorga, O., Azuaga, A. I., & van Nuland, N. A. J. (2007). The high resolution NMR structure of the third SH3 domain of CD2AP. Journal of Biomolecular NMR, 39(4), 331–336. doi:10.1007/s10858- 007-9201-7
Publications
If you're interested in applying the last NMR methods in combination with other biophysical techniques, X-Ray Crystallography and/or Electron Microscopy to membrane proteins, this is the right place.
Please contact me for further details
Members
Dr. Jose Luis Ortega-Roldan. Lecturer in Biological NMR
Dr. Lorena Varela-Alvarez. Postdoctoral research associate
Miss Alexandra Hendry. PhD student

NMR at University of Kent
The facility contains a Bruker Avance III 600 MHz NMR spectrometer with a 4-channel, 5-amplifier configuration that includes several NMR probes. Our workhorse probe is bespoke quadruple resonance inverse detection cryoprobe (QCI) with 1H, 2H 13C, 15N and 31P, plus 19F and 59Co capabilities to enable study of a large range of biomolecules. The NMR spectrometer was upgraded to its current specifications following a Wellcome Trust Equipment Award in 2010.
Research within the NMR facility covers many areas and disciplines across the School of Biosciences and through external collaborations. These include:
-
structural characterisation of biochemically and biomedically relevant biomolecules
-
investigation of the dynamic properties of proteins and peptides using NMR relaxation
-
characterisation of protein-ligand interactions
-
high resolution spectroscopy of intermediates from biosynthetic pathways
-
identification of components in crude biological samples
-
design of novel NMR and structural methodologies
-
applied computational methods and software development for both NMR and structural biology
Contact us if you'd like use the facility or discuss about how we can help your research.
Contact us
Integral membrane proteins and membrane-associated proteins are essential players in biological processes. Their functions range from transport across the membrane, cell adhesion and regulation of the cell shape. They represent around 40% of all proteins, and given their importance in the cell is not surprising that around 50% of the current drugs target membrane proteins.
We use solution NMR in combination with X-Ray crystallography and other biophysical techniques to understand the structure, interactions, dynamics and function of membrane proteins. We are also interested in NMR method development for the study of such complicated systems.
MPSF lab
